A recent review article discussed in details the current status and future prospect of protein…

Quantify PEG attached to protein or nanoparticle with the 5 validated methods you can rely on!
Protein PEGylation and Nanoparticle PEGylation – Determining the total polyethylene glycol (PEG) attached to a protein or particle, a process known as PEGylation, requires special chemical assays or biochemical tools. This is typically done to improve the substrate’s stability, solubility, or bioavailability. Below is a detailed, step-by-step explanation of methods to measure the extent of PEGylation, adhering to a formal and precise tone:
1. Understand the PEGylation Context
PEGylation involves covalently attaching PEG molecules to protein for example, typically at specific functional groups (e.g., lysine residues or N-terminal amines). The degree of PEGylation (number of PEG molecules per protein molecule) or the total mass of PEG attached must be quantified.
2. Common Methods for Quantifying PEG
Several analytical techniques can be employed to determine the total PEG attached to protein. Each method has its strengths and limitations, and the choice depends on the required precision and available equipment.
a. Spectroscopic Methods
Spectroscopic techniques are often used to detect PEGylation indirectly by measuring changes in the protein or PEG-specific signals.
- UV-Vis Spectroscopy:
- Principle: If the PEG or the linker used for conjugation has a chromophore (e.g., a fluorescent tag or a specific absorbance), UV-Vis spectroscopy can quantify the PEG by measuring absorbance at a specific wavelength. Protein itself absorbs at 280 nm due to aromatic amino acids, but PEG is typically transparent at this wavelength unless modified.
- Procedure:
- Measure the absorbance of the PEGylated protein at 280 nm to determine the protein concentration using the Beer-Lambert law.
- If the PEG or linker has a unique absorbance (e.g., at 350 nm for certain activated PEGs), measure this to estimate PEG concentration.
- Calculate the PEG-to-protein ratio by comparing concentrations, assuming the molecular weights of PEG and protein are known.
- Limitations: Requires a chromophore on the PEG or linker; non-specific absorbance may interfere.
- Fluorescence Spectroscopy:
- Principle: If the PEG is labeled with a fluorescent tag (e.g., fluorescein or rhodamine), fluorescence intensity can quantify PEGylation.
- Procedure:
- Excite the sample at the fluorophore’s excitation wavelength and measure emission intensity.
- Use a standard curve of free fluorescent PEG to quantify the amount of PEG attached.
- Normalize to protein concentration (determined via UV-Vis or other protein assays).
- Limitations: Requires fluorescently labeled PEG, which may not be used in all PEGylation protocols.
b. Chromatographic Methods
Chromatography separates PEGylated catalase from free PEG or unmodified catalase, allowing quantification.
- Size-Exclusion Chromatography (SEC):
- Principle: SEC separates molecules based on size. PEGylation increases the hydrodynamic radius of protein, causing PEGylated forms to elute earlier than unmodified catalase.
- Procedure:
- Run the PEGylated protein sample through an SEC column calibrated with molecular weight standards.
- Detect elution peaks using UV absorbance (280 nm for protein) or refractive index (for PEG).
- Quantify the area under the peaks corresponding to PEGylated protein versus free protein or PEG.
- Calculate the degree of PEGylation by comparing peak areas or using standards of known PEG-to-protein ratios.
- Limitations: Requires high-resolution columns; overlapping peaks may complicate analysis.
c. Chemical Assays for PEG Quantification
Chemical assays directly measure PEG content, often independent of protein concentration.
- Barium-Iodide Assay:
- Principle: PEG reacts with barium chloride and iodine to form a colored complex, detectable at 535 nm.
- Procedure:
- Mix the PEGylated protein sample with barium chloride and iodine solution.
- Measure absorbance at 535 nm and compare to a standard curve of known PEG concentrations.
- Subtract any background absorbance from free PEG or protein (run controls with non-PEGylated protein).
- Calculate total PEG mass and divide by protein mass (determined separately) to estimate the PEG-to-protein ratio.
- Limitations: Sensitivity depends on PEG molecular weight; requires careful control experiments.
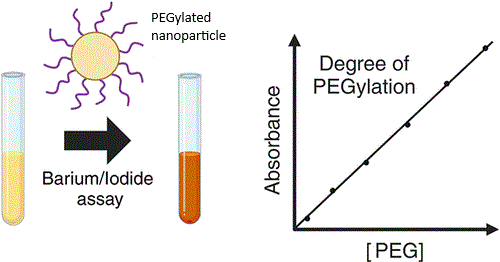
Barium-Iodide Assay for colorimetric quantification of PEG
Read more at Biomacromolecules 2025, 26, 6, 3689–3699
- TNBS Assay for Free Amines:
- Principle: PEGylation often targets lysine residues, reducing the number of free amines. Trinitrobenzenesulfonic acid (TNBS) reacts with free amines to produce a colored product, allowing indirect quantification of PEGylation.
- Procedure:
- React PEGylated and non-PEGylated protein with TNBS and measure absorbance at 420 nm.
- Compare the reduction in free amines in the PEGylated sample to the non-PEGylated control to estimate the number of PEG molecules attached per protein molecule.
- Limitations: Only applicable for amine-targeted PEGylation; requires knowledge of protein’s amine content.
d. Mass Spectrometry
- Principle: Matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization (ESI) mass spectrometry can determine the molecular weight of PEGylated protein, revealing the number of PEG molecules attached.
- Procedure:
- Purify the PEGylated protein to remove free PEG and other contaminants.
- Analyze the sample using MALDI-TOF or ESI-MS to obtain the molecular weight.
- Compare the observed molecular weight to that of unmodified protein. The mass increase corresponds to the total PEG attached (each PEG molecule adds a known molecular weight, e.g., 5 kDa for a 5,000 Da PEG).
- Calculate the number of PEG molecules per protein.
- Limitations: Requires high-resolution instrumentation; polydispersity of PEG may complicate analysis.
e. SDS-PAGE Analysis
- Principle: Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) separates proteins by size. PEGylation increases the apparent molecular weight of protein, causing a band shift.
- Procedure:
- Run PEGylated and non-PEGylated protein on an SDS-PAGE gel.
- Stain the gel with Coomassie Blue for protein or barium iodide for PEG to visualize bands.
- Estimate the degree of PEGylation by comparing band shifts to molecular weight standards or known PEGylated controls.
- Notes: Semi-quantitative; PEG can affect staining efficiency, requiring careful interpretation. This is a general method for detecting pegylated proteins directly after SDS-PAGE. The proteins to which polyethylene glycol (PEG) molecules are attached are stained with a barium iodide solution. The staining is based on the formation of a barium iodide complex with PEG. The described method combines a specific staining of PEG molecules with the high resolution of the SDS-PAGE method. It is shown that pegylated protein is detectable on SDS-PAGE as well as on IEF at concentrations that are not detectable by Coomassie protein staining. The determination of the molecular weight of pegylated protein by calibrating SDS-PAGE with polyethylene glycol of different molecular weight is feasible. Under standard gel conditions, PEG showed linear mobility during electrophoresis. However, the use of non-pegylated proteins as standards is improper due to the lower mobility of the pegylated protein during electrophoresis.
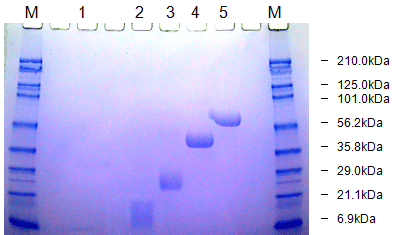
Detection and molecular weight determination of polyethylene glycol-modified protein by staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Read more at 10.1016/0003-2697(92)90460-O, and 10.1016/0026-265X(75)90038-7
3. Practical Considerations
- Purification: Before analysis, purify the PEGylated protein using dialysis, ultrafiltration, or chromatography to remove free PEG, which can interfere with quantification.
- Controls and Standards: Use non-PEGylated protein and free PEG as controls. Prepare standard curves for PEG quantification (e.g., in barium-iodide or fluorescence assays).
- PEG Molecular Weight: Know the molecular weight of the PEG used (e.g., 5 kDa, 20 kDa) to interpret mass increases or chromatographic shifts.
- Multiple PEGylation Sites: Protein may have multiple lysine residues, so PEGylation may result in a heterogeneous mixture (mono-, di-, or multi-PEGylated forms). Techniques like SEC or mass spectrometry can resolve these species.
- Enzyme Activity: Verify that PEGylation does not significantly impair the enzyme’s activity, as this may affect downstream applications if the protein is an enzyme.
4. Example Calculation
Suppose mass spectrometry shows a PEGylated protein molecular weight of 250 kDa, while unmodified protein is 240 kDa, and the PEG used is 5 kDa:
- Mass increase = 250 kDa – 240 kDa = 10 kDa.
- Number of PEG molecules = 10 kDa / 5 kDa = 2 PEGs per protein molecule.
- Total PEG mass = 10 kDa per protein molecule.
5. Conclusion
To determine the total PEG attached to protein, a combination of purification (e.g., SEC) and analytical techniques (e.g., mass spectrometry, barium-iodide assay, or TNBS assay) provides the most reliable results. Select methods based on available equipment, the type of PEG used, and the desired precision. Ensure proper controls and standards are included to account for background signals and confirm the accuracy of the quantification.
If further details are needed (e.g., specific protocols, reagent concentrations, or instrument settings), please contact technical support at Creative PEGWorks.
Related Posts
- The Case for Protein PEGylation
- Fluorescent or Radioactive PEG-like Nanoprobes
Researchers from the Massachusetts General Hospital and Harvard Medical School developed the novel, "PEG-like Nanoprobes,’’…
- PEGylation Protocols and Literature References
Creative PEGWorks has collected hundreds of papers published in peer-reviewed journals citing the use of…
